Kelp and their microbiome: ongoing research in the lab
We are isolating and sequencing the genomes of bacterial taxa that are associated with kelp. We are discovering the metabolisms of kelp-associated bacteria through analysis of their genomes and laboratory assays of bacterial metabolisms and nutrient use. Our studies extend over a broad geographic range from healthy kelp beds to those in decline.
Thanks to NSF-OCE2329475 for support & collaborators Sam Light & Jake Waldbauer.


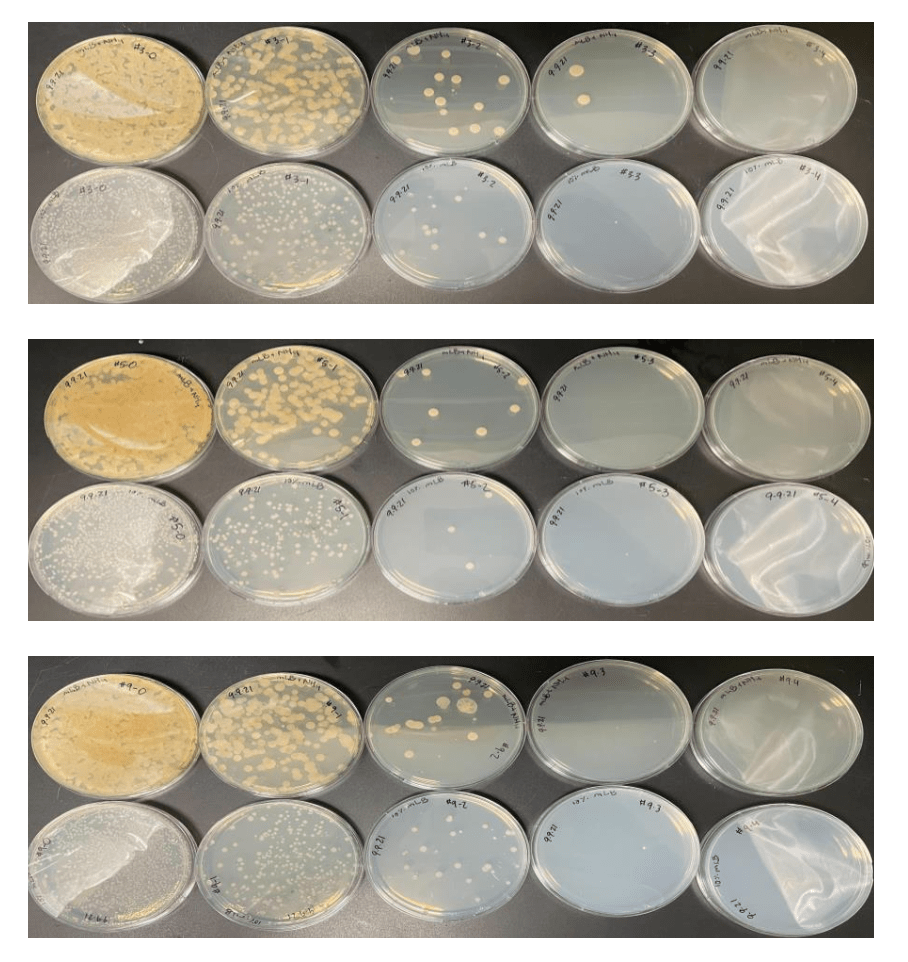
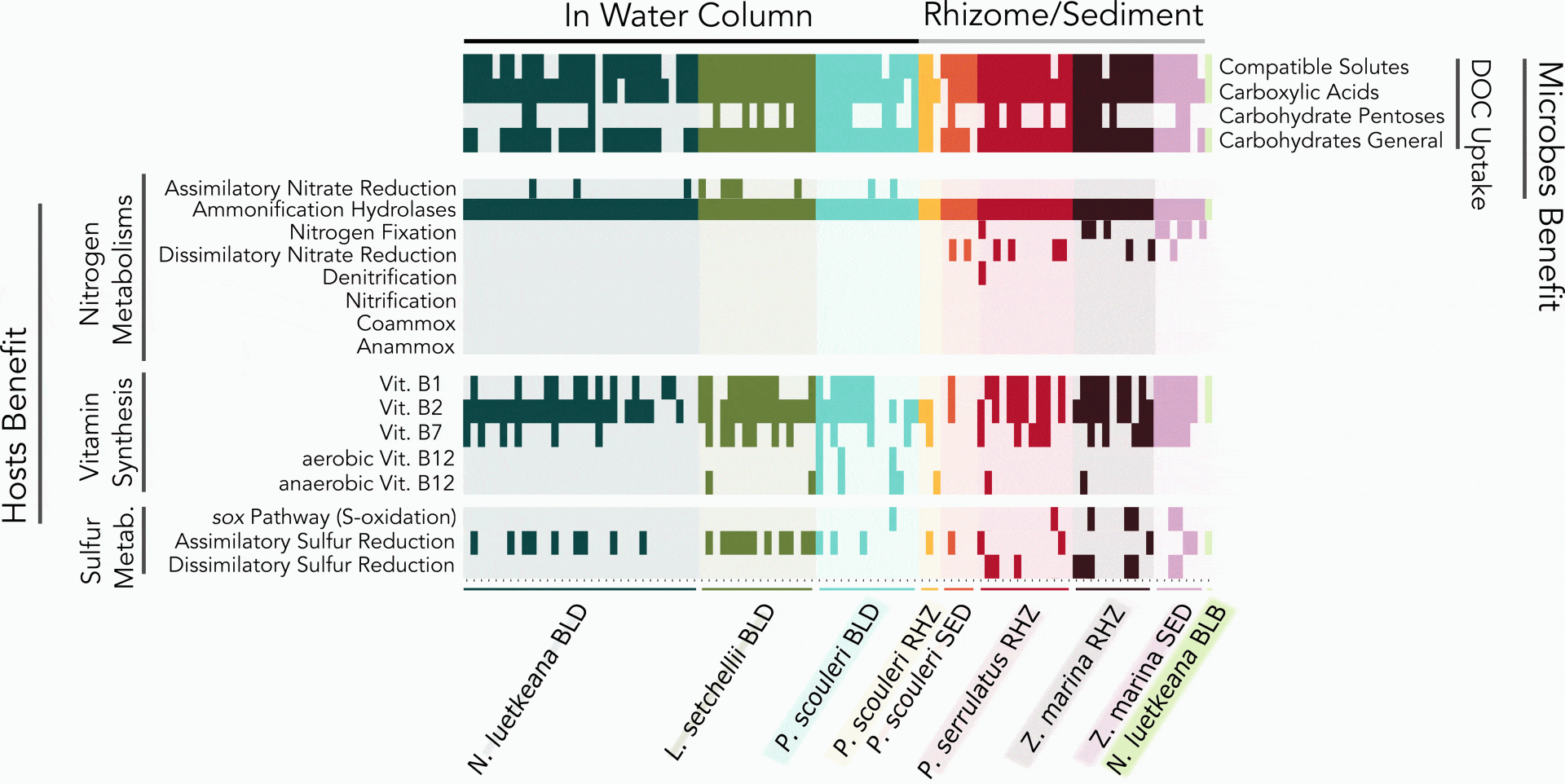
The bull kelp in Washington state host a diverse microbiome. We have isolated and fully sequenced the genome of 16 of these bacteria. Further, we show that the amino acid metabolism of these bacteria provide ammonium. They also use the amino acids that are abundant in seawater. Their genome suggests metabolisms including vitamin production and response to reactive oxygen stress.
Kelp host a diverse microbial community and microbes have metabolisms that include vitamin production and nutrient recycling, revealed in their genomes.
Kelp beds in Washington state change local seawater chemistry and entrain a different microbial assemblage in their vicinity. See:
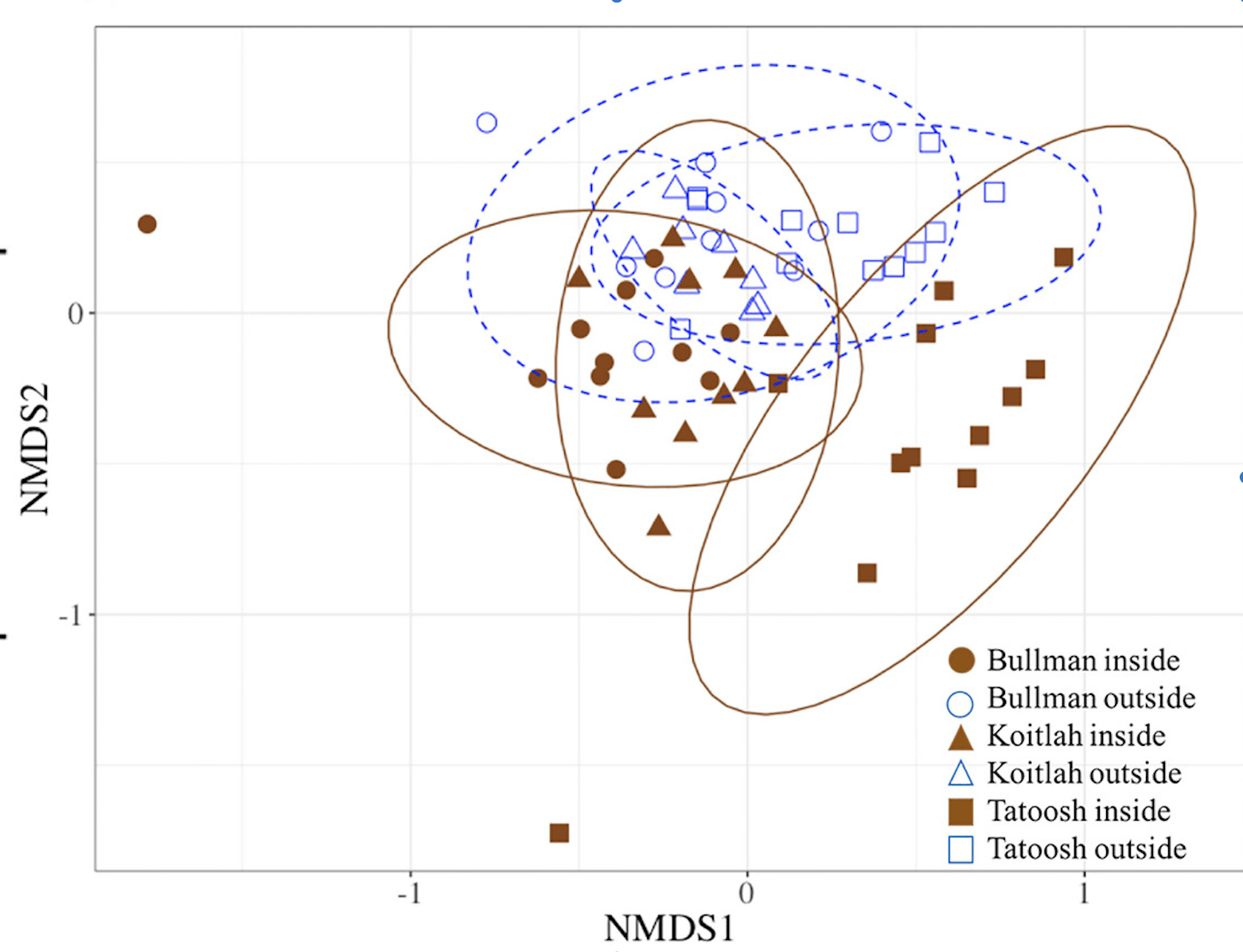

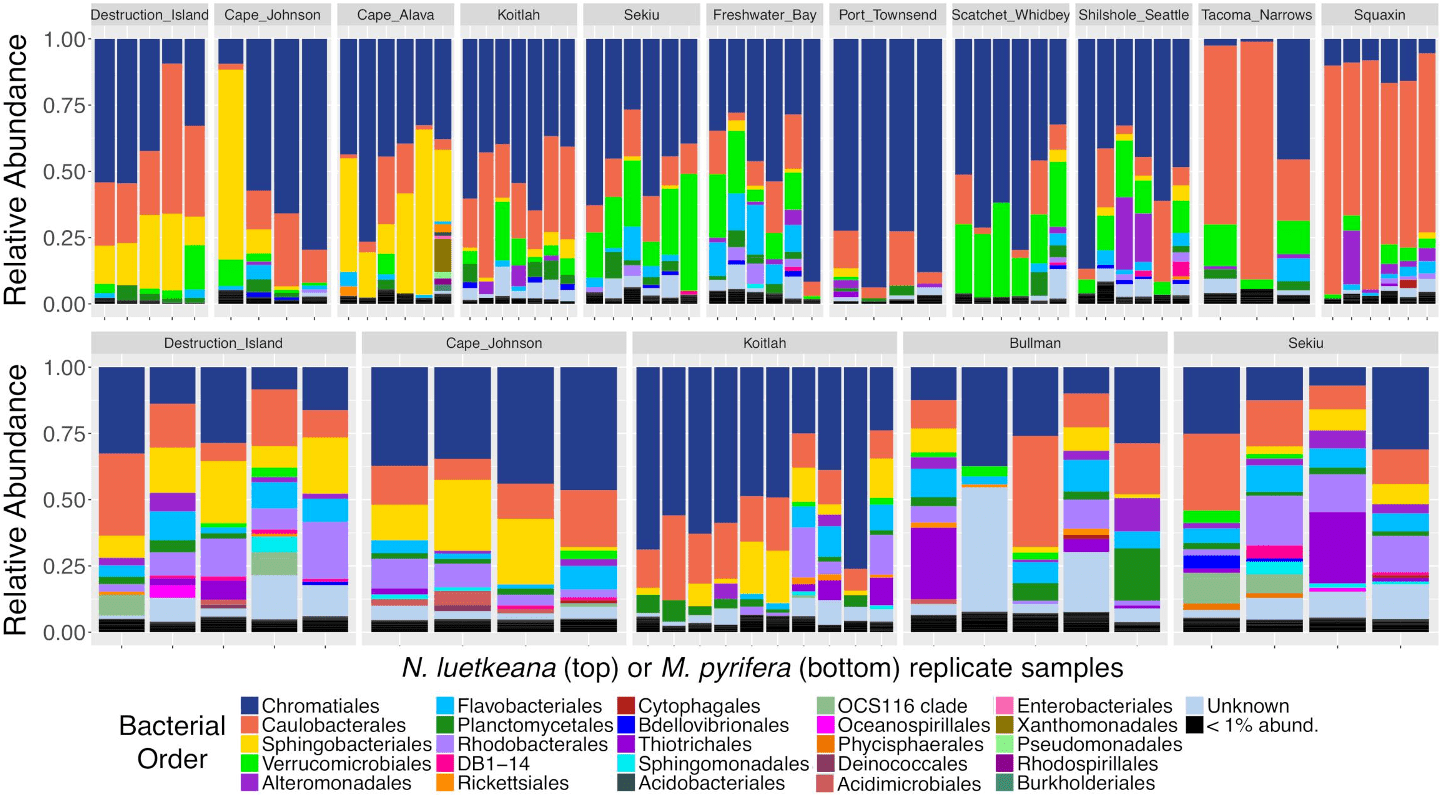
Brooke Weigel‘s study of the microbiome on bull kelp and giant kelp in Washington state reveal that canopy kelp have unique taxa associated with them and the taxonomic composition differs geographically.
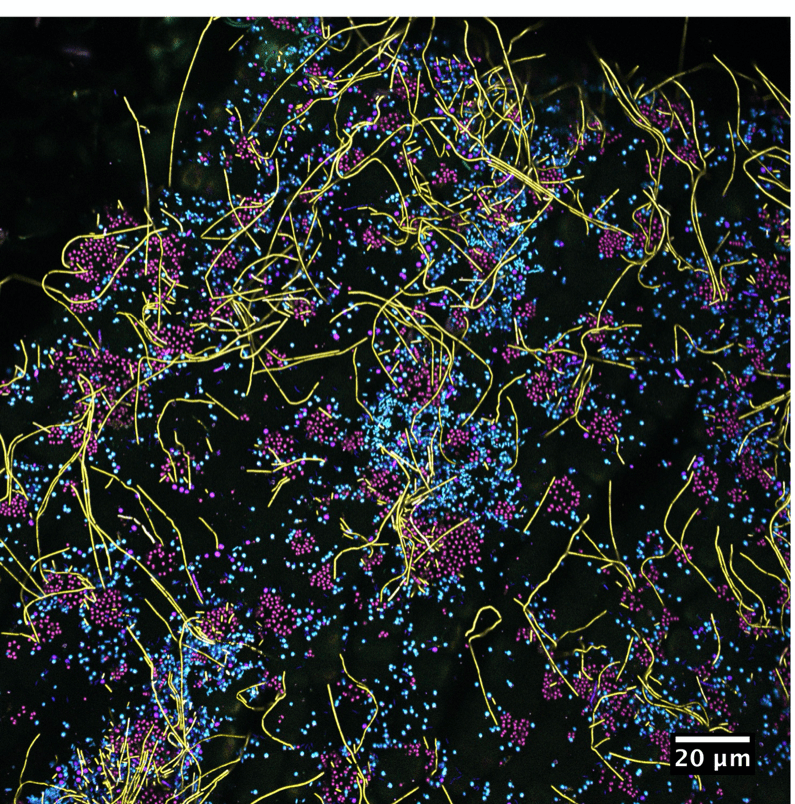
Incredible imaging by colleagues at the Marine Biological Lab at Woods Hole reveal a complex bacterial community on the surface of kelp.
The taxonomic diversity of bacteria on kelp includes a richer set of microbial species in areas where kelp are healthy and thriving. Imaging by Tabita Ramirez-Puebla and Jessica Mark Welch show this with their CLASI-FISH methodology and contrast outer coast kelp with Puget Sound kelp in Washington state.
Metagenomics reveals that the taxonomic and functional diversity of bacteria living on the bull kelp is diverse.


